About
Introduction of RICAtlas
RIC-Atlas is a resource-based database of RNA-RNA interactions. RNA molecules usually interact with each other, to regulate critical biological processes. So, understanding the mechanisms and functions of RNA-RNA interactions is crucial.
RNA in situ conformation sequencing (RIC-seq) is a technique for capturing protein-mediated RNA-RNA proximal interactions globally in living cells at single-base resolution, which described the interactions between RNAs more precisely.
The RIC-Atlas currently identified 10 human cell lines and obtained 820,086 high-confidence enhancer-promoter interactions.
RIC-seq workflow
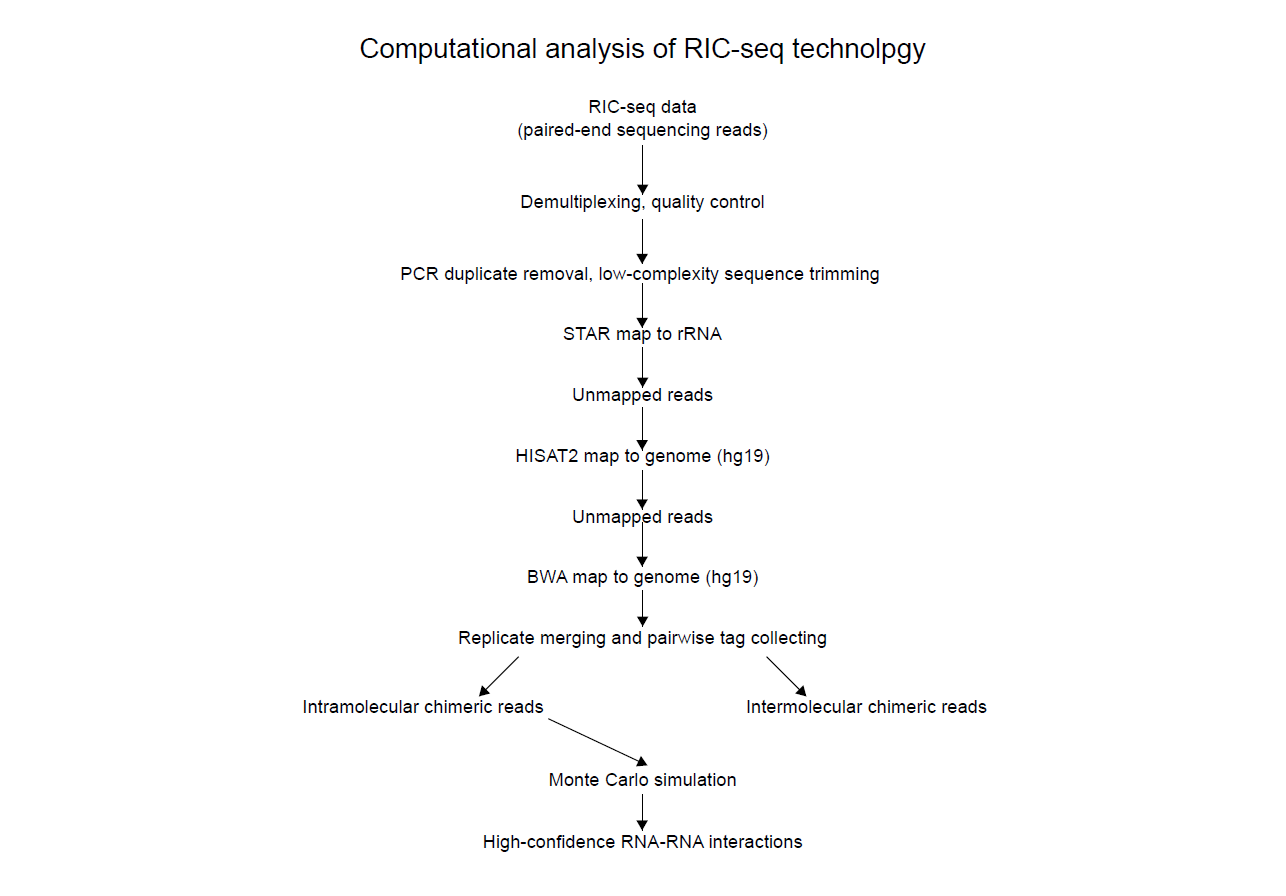
RIC-seq libraries were paired-end-sequenced at Novo gene.
The analysis strategy is illustrated above. Adapters were trimmed off with the Trimmomatic program (v.0.36)36, and PCR duplicates were removed by homemade scripts.
After filtering, the paired reads were first aligned to 45S pre-rRNA, and the remaining reads were mapped to the human reference genome (assembly version: hg19) using the STAR software.
The STAR mapping produced normally mapped reads (Aligned_out.sam) and chiastically mapped reads (Chimeric_out.sam). The low-quality and secondary mapping results were filtered using the SAMtools package.
Definition of enhancer and promoter
Promoters were defined as the H3K4me3 ChIP–seq peak-enriched regions within ±5 kb of the transcription start sites.
To define enhancers, we first removed the H3K27ac ChIP–seq peaks within ±2.5 kb of known gene promoters and then stitched the remaining H3K27ac peaks clustered within a 12.5-kb region together and classified them as enhancers.
Super-enhancers were defined based on the coverage of H3K27ac ChIP–seq signals using a Rank Ordering of Super-Enhancers (ROSE) algorithm. EPRIs were identified as pairwise interacting RNA fragments transcribed from those enhancer and promoter regions.
Obtain high-confidence E-P interactions
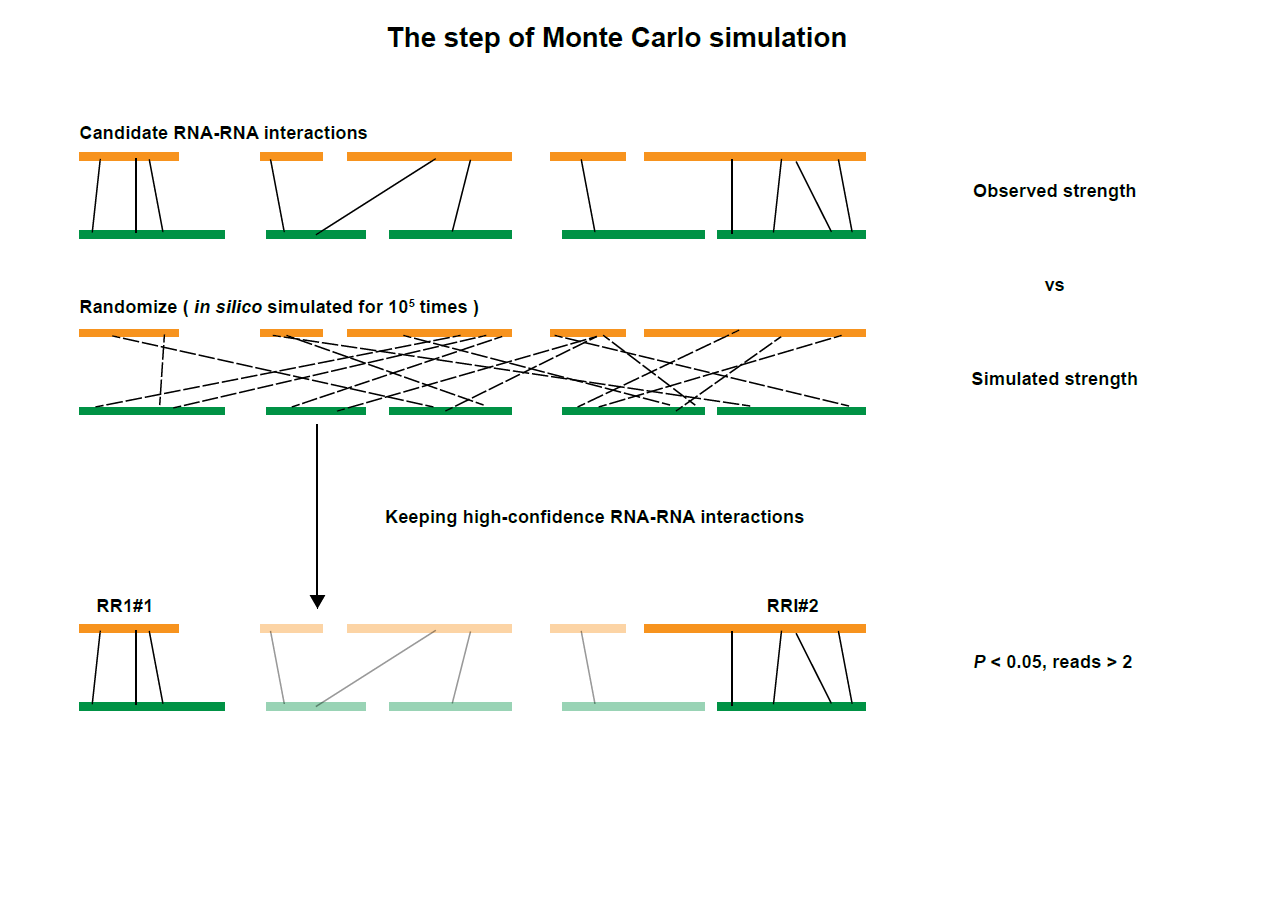
After obtaining Enhancer-Promoter interactions, we adopted a previously described Monte Carlo simulation to detect significant contacts by comparing observed pairwise interactions to the counts of random ligations, in which 100,000 simulations were performed. Enhancer-Promoter interactions with local-background-corrected P values lower than 0.05 were used in this database.
The figure above showed the diagram of strategies for defining enhancer and promoter regions, assigning chimeric reads to enhancer-promoter pairs, and identifying high-confidence Enhancer-Promoter RNA interactions.
Citations
Cai, Z. et al. RIC-seq for global in situ profiling of RNA–RNA spatial interactions. Nature 582, 432–437 (2020). https://doi.org/10.1038/s41586-020-2249-1
Liang, L., Cao, C., Ji, L. et al. Complementary Alu sequences mediate enhancer–promoter selectivity. Nature 619, 868–875 (2023). https://doi.org/10.1038/s41586-023-06323-x